Plant membrane signal transduction
Cells are surrounded by a lipid bilayer, the plasma-membrane. Embedded in this membrane are receptor proteins that serve as the cell’s eyes and ears, helping a cell to communicate with the outside world and with other cells. All receptor proteins have to accomplish three basic tasks in the communication process; the specific recognition of an outside signal , the transport of the signal across the membrane, and the initiation of a response to that signal in the cell’s interior. These steps are collectively known as trans-membrane signaling.
Different cells use different receptor proteins to mediate trans-membrane signaling. The key players in bacteria are sensor histidine kinases (see below). Major membrane signaling receptors in animals are G-protein coupled receptors and receptor tyrosine or serine/threonine kinases. These receptor families have been systematically studied both in cells and in isolation. Much less is known however about trans-membrane signaling in plants. While plants and animals are both highly evolved multicellular organisms and share many biosynthetic and signaling pathways, membrane signaling in plants is remarkably distinct from the animal world.
Plants do neither contain G-protein coupled receptors nor canonical tyrosine receptor kinases, but instead rely on sensor histidine kinases (which they probably adopted from bacteria) and on a family of plant-unique receptor kinases (they are commonly referred to as receptor-like kinases, but we know now that many family members are indeed both kinases AND receptors).
Leucine-rich repeat receptor kinases
Several hundred of these receptor kinases exist in the small genome of the model plant Arabidopsis, and the majority of them shares a common architecture: a N-terminal extracellular ligand binding domain composed of leucine-rich repeats (LRRs), a single membrane spanning helix, and a cytoplasmic kinase domain. LRR receptor kinases have diverse functions in plants, and for example regulate plant growth, development and the interaction with the environment. Their corresponding ligands range from small molecules to peptides and entire proteins. Forward genetic screens in Arabidopsis have identified cellular functions for many of these receptors, and different receptor-ligand pairs have been described. We study how these receptors bind their ligands, how they transduce the signal across the membrane and how they activate cytoplasmic signaling cascades.
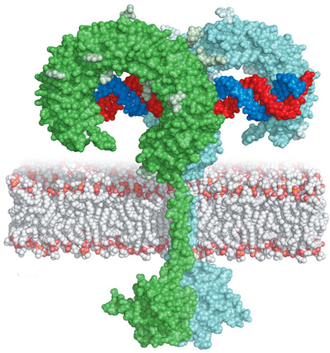
The architecture of plant LRR receptor kinases is reminiscent of animal Toll-like innate immunity receptors (TLRs). In TLR signaling, ligands glue the horseshoe-shaped domains of two neighboring receptors together to initiate the cellular response; a process called ligand-induced homodimersation (Figure 1). It was thus reasonable to assume that the extracellular ligand binding domain of plant LRR receptor kinases would also form a TLR-like horseshoe structure that binds its ligand along a dimer interface (Figure 1). Surprisingly, this assumption turned out to be incorrect (see below).
Steroid hormone sensing by the plant receptor kinase BRI1
Steroid hormones are regulators of body shape and size in higher organisms. Animal steroid receptors reside in the nucleus ( nuclear hormone receptors ), but in plants the membrane LRR receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) acts as a direct receptor for brassinosteroids . To understand how BRI1 can recognize its steroid ligand at the plasma membrane, we determined the crystal structure of the entire extracellular ligand binding domain of BRI1 (a 120 kDa glycoprotein) in the pre- and absence of brassinolide, the most potent brassinosteroid in Arabidopsis.
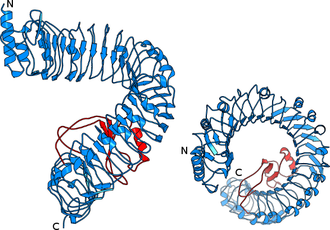
Instead of having the expected horseshoe structure, the leucine-rich repeats in BRI1 form a beautiful superhelix composed of 25 LRRs, completing slightly more than one full turn (Figure 2). The sequence pattern that causes BRI1 to twist into a superhelix is also found in other plant receptor kinases, and thus these receptors also contain twisted or superhelical ligand binding domains (see below, Hohmann, Lau & Hothorn, Annual Reviews of Plant Biology, 2017).
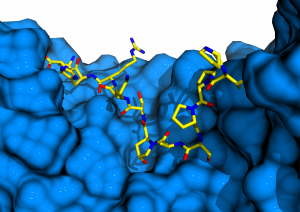
In contrast to previously described LRR proteins, BRI1 is able to recognize a small hydrophobic ligand. A unique adaptation to this challenge is the insertion of a small folded island domain into the regular repeat structure (Figure 3).

This island domain is largely disordered in the unliganded receptor structure. Steroid binding induces a conformational rearrangement and fixing of the island domain. We speculated back in 2011 (Hothorn et al., Nature, 2011) that the steroid ligand and the now fully ordered island domain together may represent a docking platform for a shape-complementary helper protein that would allow the receptor to become activated. Candidates for such helper proteins were the small somatic embryogenesis receptor kinases (SERKs), which had been implicated in brassinosteroid signalling (Li et al., Cell, 2002, Nam & Li, Cell, 2002). We thus set out to study, whether SERK proteins are the envisioned shape-complementary helpers required to activate BRI1 signal transduction.
Plant steroid receptor activation by somatic embryogenesis co-receptor kinases
Postdoctoral fellow Julia Santiago and lab manager Christine Henzler managed to express the isolated LRR ectodomains of BRI1 and of the putative co-receptor kinase SERK1 in insect cells.
While these domains do not physically interact in the absence of the steroid hormone, they form very tight heterodimers in the presence of even minute amounts of brassinolide, the most potent brassinosteroid in Arabidopsis (Santiago, Henzler and Hothorn, Science, 2013).
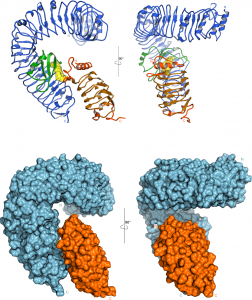
A crystal structure of the BRI1-brassinolide-SERK1 complex revealed that the hormone acts as a molecular glue, which promotes association of the LRR domains of receptor and co-receptor (Figure 4).
The hormone binding pocket is formed by residues from both the receptor and from the co-receptor, and thus both proteins are required for high-affinity sensing of the steroid molecule.
Importantly, the steroid-promoted association of the BRI1 and SERK1 LRR domains brings their C-termini (which connect to the trans-membrane helices and to the cytoplasmic kinase domains) in close proximity. We thus assume that steroid binding at the cell surface to BRI1 and SERK1 allows their kinase domains in the cytoplasm to interact. The movie below depicts how the extracellular BRI1 LRR domain (in blue) binds brassinolide (in yellow) using its island domain (in magenta). Brassinolide binding allows BRI1 to interact with the LRR domain of the co-receptor kinase SERK1 (in orange). The green spheres at the end of the animation depict the location of known missense alleles in BRI1 and BAK1, which affect brassinosteroid signaling (movie by Kelvin Lau, generated using the program Chimera).
The kinase domains of receptor and co-receptor can interact and transphosphorylate each other
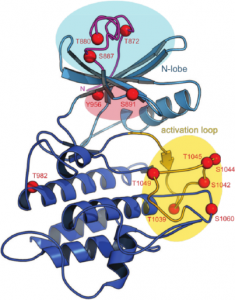
We have recently begun to dissect the cytoplasmic side of signaling (Bojar et al., Plant J, 2014). Undergraduate researcher Daniel Bojar found that the kinase domain of BRI1 is structurally related to the Pelle/IRAK family of kinases in animals, which rationalizes why BRI1 is a dual specificity kinase. The auto-phosphorylation pattern of BRI1 is very complex (Figure 5) and we could find BRI1 homodimers as well as heterodimers with SERK kinase domains. Our interpretation is that BRI1, SERKs and other plant LRR kinases may form homodimers trough their kinase domains as a basal or resting state, and may heterodimerise with a co-receptor kinase domain for activation (Fig. 6). Importantly, trans-phosphorylation between BRI1 and SERKs has been demonstrated in vivo (Wang et al., Dev Cell, 2008). We mapped one interaction side of BRI1 with SERKs to the C-lobe of the BRI1 kinase domain and thus an asymmetric arrangement in the heterodimer is possible, with one kinase acting as the enzyme and the partner acting as substrate. Interestingly, the BRI1 inhibitor protein BKI1 also binds to the C-lobe of BRI1 and acts by inhibiting formation of the BRI1-SERK kinase heteromer (Jaillais et al., Genes Dev, 2011).
A complete biochemical and genetic validation of the BRI1 activation mechanism by PhD student Ulrich Hohmann revealed that the extracellular LRR domains of BRI1 and SERKs only interact in the presence of the brassinolide ligand, and that a receptor – co-receptor heterodimer is required for high affinity ligand binding and receptor activation. Signaling specificity is encoded in kinase domain of the receptor, not in the kinase domain of the co-receptor (Hohmann et al., PNAS, 2018).
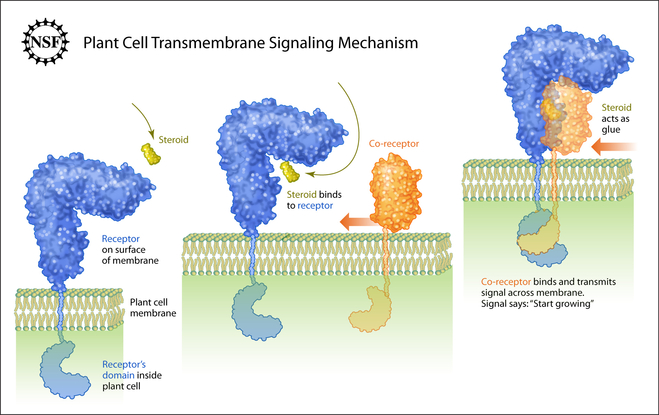
Shedding of floral organs requires the receptor kinase HAESA, the peptide hormone IDA and the co-receptor kinase SERK1
Have you ever wondered how trees in the fall know when to let go? These abscission processes (shedding of old or no longer required organs) are controlled by the plant receptor kinase HAESA and its cognate peptide hormone ligand IDA. Together with our collaborators Melinka Butenko and Mari Wildhagen from the University of Oslo, Norway, postdoctoral fellows Julia Santiago and Benjamin Brandt could demonstrate that HAESA directly senses the IDA peptide, specifically an active 12mer peptide carrying a central hydroxyproline residue (a post-translational modification common to many plant peptide hormones) (Figure 7). However, HAESA by itself cannot bind IDA with high affinity. It requires a co-receptor kinase, which like in the case of BRI1 (see above), completes the ligand binding pocket. The HAESA co-receptor turned out to be SERK1, the receptor kinase which also BRI1 requires for receptor activation (Santiago et al., eLife, 2016). The movie shows the LRR domain of HAESA (in blue) interacting with the active peptide hormone IDA (in yellow). The co-receptor kinase SERK1 (in orange) binds next, specifically interacting with the IDA C-terminus. The zoom in highlights the C-terminal 3 amino-acids in IDA as well as the central hydroxyproline, which anchors the peptide to the receptor. (movie by Kelvin Lau, using the program Chimera).
The receptor activation mechanism for the steroid receptor BRI1 and the peptide hormone receptor HAESA are highly conserved, as chimera between both receptors are functional in planta (Hohmann et al., PNAS, 2018).
The LRR receptor kinases GSO1/GSO2 sense sequence divergent CIF peptides to control barrier formation in different plant organs.
Plants contain many different classes of peptide hormones of different length, sequence, and carrying different post-translational modifications. In collaboration with the lab of Niko Geldner at the University of Lausanne, postdoctoral fellow Satohiro Okuda studied the receptor kinases GSO1 and GSO2 involved in plant embryo development and in the formation of the Casparian strip, a nutrient diffusion barrier in the root. Satohiro solved the structure of the huge GSO1 LRR ectodomain and went on to show that GSO1 and GSO2 can bind a newly discovered family of sequence diverse tyrosine sulfated peptide hormones (Okuda et al., PNAS, 2020; Doll et al., Science, 2020) (Figure 8).
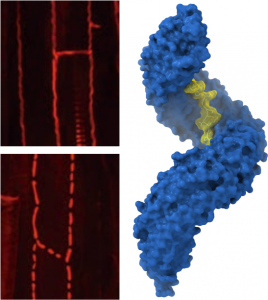
The interaction between GSO1 and CIFs is much more tight than previously reported for other plant peptide hormone – receptor pairs and the post-translational modification of the peptide is a critical determinant for binding specificity. Like BRI1 and HAESA, also GSO1 and GSO2 use SERK co-receptor kinases for receptor activation (Okuda et al., PNAS, 2020; Hohmann et al., Plant Cell, 2020). Thus, a large family of LRR receptor kinases exists in plants, with each receptors binding a specific (family of) ligand(s) but all sharing the same receptor activation mechanism. A more detailed description of other SERK-dependent receptor systems can be found in our review by Hohmann, Lau & Hothorn, Annual Reviews of Plant Biology, 2017.
Negative regulation of SERK-dependent receptor kinases by BIR receptor pseudokinases.
Receptor activation of many plant LRR receptor kinases relies on the ligand-dependent heterodimerization with shape-complementary SERK co-receptor kinases (Hohmann, Lau & Hothorn, Annual Reviews of Plant Biology, 2017). By studying the previously characterized elongated gain-of-function allele in the co-receptor kinase SERK3, we found that these receptor – ligand – co-receptor complexes can be negatively regulated by a family of BIR receptor pseudokinases. PhD student Ulrich Hohmann uncovered that the LRR ectodomains can consitutively bind the LRR domains of SERK co-receptors, thereby blocking their interaction with ligand-bound LRR receptor kinases (Hohmann et al., Nature Plants, 2018) (Figure 9).
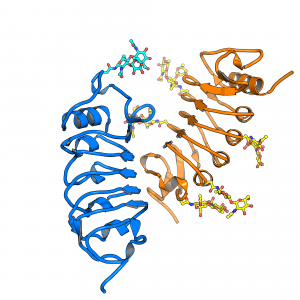
Ulrich exploited his finding that BIR LRR domains can bind SERKs in the absence of any ligand to create various BIR chimera that allow for the consitutive activation of LRR receptor kinase pathways in plants (Hohmann et al., Plant Cell, 2020, collaboration with Martin Bayer, MPI Dev Biol, Tuebingen and Marie Barberon, Uni Geneva). These experiments define BIR proteins as general negative regulators of SERK-dependent LRR receptor kinases and again confirm the receptor activation mechanism outlined above. It is of note though, that not all LRR receptor kinases appear to depend on SERKs, other co-rececptor and/or activation mechanisms may exist (Hazak et al., EMBO Rep, 2017; Anne et al., Development, 2018, collaboration with Chrisitian Hardtke, Uni Lausanne). Also, not all signaling complexes may be heterodimeric, as modular receptors systems exist (for example see Hohmann et al., Acta D, 2019).
Receptor kinases with non-LRR ectodomains
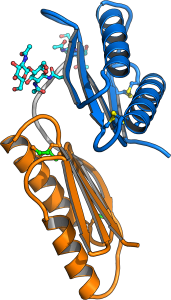
hile LRR receptor kinases represent the largest group of membrane receptor kinases in plants, many other receptors with unique ectodomain architectures have evolved in the green lineage. Postdoctoral fellow Benjamin Brandt resolved the ectodomain structures of the cysteine-rich receptor kinases previously termed DUF26 domains. He found these ectodomains to form a unique tandem arrangement of malectin domains, carbohydrate binding modules previously characterized in animals (et al., Commun Biol., 2019) (Figure 10).
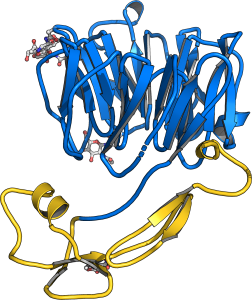
Postdoctoral fellow Satohiro Okuda studied the architecture and physiological function of plant-unique CRINKLY4 receptor kinases. By solving structures of the receptors ACR4 from Arabidopsis and PpCR4 from the moss Physcomitrella patens he uncovered that CRINKLY4 receptor kinase ectdomains harbor an N-terminal WD40 and a C-terminal cysteiner-rich domain with weak structural homology to tumor necrosis factor receptors (Okuda et al., bioRxiv, 2020) (Figure 11). The entire ectodomain is stablized by many disulfide bonds. The C-terminal cysteine rich domain and the kinase activity of the receptor are dispensible for function and the moss receptor can functionally replace the Arabidopsis receptor in reverse genetic experiments. A large ligand bindin groove in the WD40 domain suggests that unique ligands may exist for CRINKLY4 receptor kinases.
Sensor histidine kinase hormone receptors in plants
Plants do not only rely on receptor kinases to mediate transmembrane signaling, but also on sensor histidine kinases . These receptor proteins are normally found in prokaryotic genomes and can be considered the key players in bacterial membrane signal transduction.
Arabidopsis contains several sensor histidine kinase proteins, including a putative osmosensor, a family of hormone receptors that recognize the gaseous plant hormone ethylene , and a set of three cytokinin receptors. Cytokinins are classic phytohormones derived from adenine . Cytokinins control diverse processes in plant growth and development, and are essential to maintain plant stem cell populations. Cytokinin receptors are eukaryotic sensor histidine kinases. These receptors bind the hormone with their extracellular sensor domain and initiate a classical two-component phosphorelay in the cytoplasm. By solving the crystal structure of the plant cytokinin receptor we found, that despite being divergent in sequence, plant and bacterial histidine kinases are very similar (Figure 12) (Hothorn et al., Nature Chem Biol, 2011).
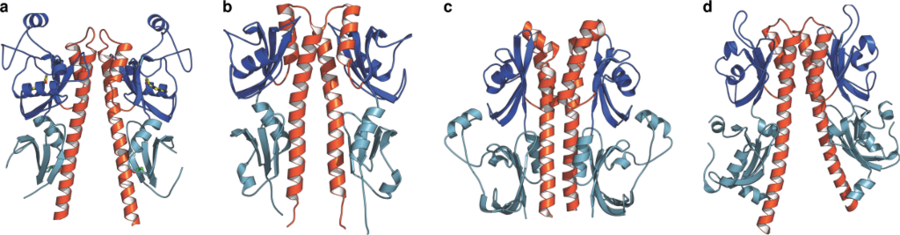
Cytokinin receptors can sense chemcially diverse hormone ligands
Cytokinin receptors can not only respond to a diverse set of naturally occurring adenine-type cytokinins, but also bind synthetic urea-type compounds. These molecules are so diverse in chemical structure that it is hard to imagine how the receptor gets the job done. We thus crystallized the extracellular sensor domain of the Arabidopsis cytokinin receptor AHK4 in complex with many different natural and synthetic cytokinins, and compared them side-by-side. Figure 13 shows that the receptor contains a binding pocket that can accommodate very different ligands, by simply checking a few critical contacts on one hand, and by providing a simple size-selectivity filter on the other hand. In this way, AHK4 can interact with diverse adenine-type ligands (Figure 14).
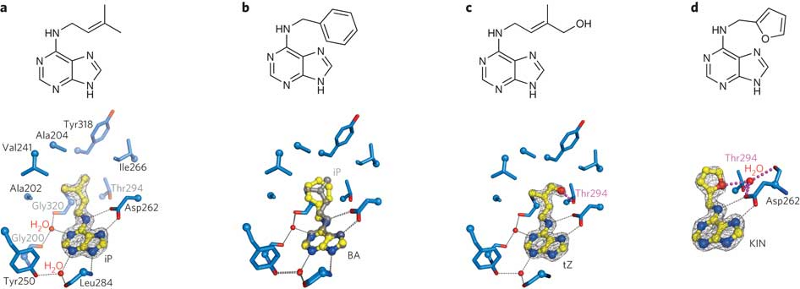
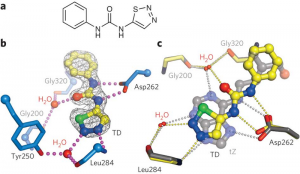
Although synthetic urea-type cytokinins look very different from the naturally occurring hormones, we found that they do mimic the critical contacts, basically hijacking the binding site (Figure 14). Synthetic cytokinins are important growth regulators, herbicides and defoliants in agriculture. Thus, on the more practical side, the different AHK4 structures suggest how to design new cytokinins rationally, with potential application in basic research and in the field.
