A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis
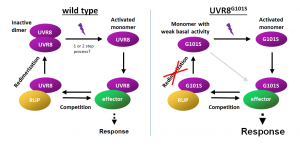 Coping with UV-B is crucial for plant survival in sunlight. The UV-B photoreceptor UVR8 regulates gene expression associated with photomorphogenesis, acclimation and UV-B stress tolerance. UV-B photon reception by UVR8 homodimers results in monomerization, followed by interaction with the key signaling protein COP1. Roman Ulm’s group, in collaboration with Michael Hothorn‘s group, has discovered a UV-B hypersensitive UVR8 photoreceptor (UVR8G101S) that confers strongly enhanced UV-B tolerance and generated a novel UVR8 variant based on the underlying mutation that shows extremely enhanced constitutive signaling activity. These findings provide key mechanistic insight into how plants respond and acclimate to UV-B radiation.
Coping with UV-B is crucial for plant survival in sunlight. The UV-B photoreceptor UVR8 regulates gene expression associated with photomorphogenesis, acclimation and UV-B stress tolerance. UV-B photon reception by UVR8 homodimers results in monomerization, followed by interaction with the key signaling protein COP1. Roman Ulm’s group, in collaboration with Michael Hothorn‘s group, has discovered a UV-B hypersensitive UVR8 photoreceptor (UVR8G101S) that confers strongly enhanced UV-B tolerance and generated a novel UVR8 variant based on the underlying mutation that shows extremely enhanced constitutive signaling activity. These findings provide key mechanistic insight into how plants respond and acclimate to UV-B radiation.
This article was published in PNAS on February 9, 2021.
Discovery of a phosphate sensing and signaling pathway in plants
Cells require sufficient amounts of the element phosphorus to build their membranes and to store and copy genetic information. Phosphorus is taken up by cells in the form of inorganic phosphate, an important signaling molecule and energy currency. While we take up sufficient amounts of phosphate with our diet, plants have to mobilize and take up phosphate from the soil, where it is poorly bioavailable. Phosphate thus limits the growth of plants, and phosphate fertilizers have to be used to maximize crop yields. How plant cells measure cellular phosphate levels and how they decide if and when to take up more phosphate is poorly understood.
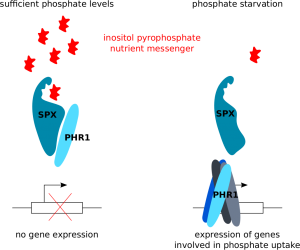 The Hothorn lab has previously shown that phosphate-rich inositol pyrophosphates are nutrient messengers in plants and identified SPX domains as their cellular receptors. In a new report, the Hothorn, Hiller (Biozentrum Basel) and Fiedler (FMP Berlin) labs now report that inositol pyrophosphates control the activity of the transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1). When there is enough phosphate in the cell, inositol pyrophosphates bind to the SPX receptor which in turn binds to PHR1, keeping it in a isolated form unable to active gene expression. When phosphate becomes limiting, inositol pyrophosphates are less abundant, the SPX – PHR1 complex dissociates and the free transcription factor can interact with itself and activate the expression of genes involved in phosphate uptake. This signaling mechanism may now be exploited towards the development of phosphate starvation tolerant crops that would require less phosphate fertilizer.
The Hothorn lab has previously shown that phosphate-rich inositol pyrophosphates are nutrient messengers in plants and identified SPX domains as their cellular receptors. In a new report, the Hothorn, Hiller (Biozentrum Basel) and Fiedler (FMP Berlin) labs now report that inositol pyrophosphates control the activity of the transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1). When there is enough phosphate in the cell, inositol pyrophosphates bind to the SPX receptor which in turn binds to PHR1, keeping it in a isolated form unable to active gene expression. When phosphate becomes limiting, inositol pyrophosphates are less abundant, the SPX – PHR1 complex dissociates and the free transcription factor can interact with itself and activate the expression of genes involved in phosphate uptake. This signaling mechanism may now be exploited towards the development of phosphate starvation tolerant crops that would require less phosphate fertilizer.
The article was published in Nature Communications, on January 15th 2021.
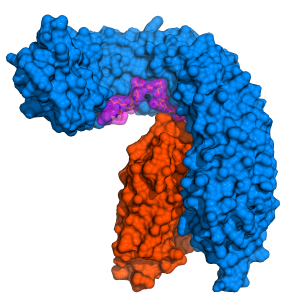
Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2
UNIGE press release, Jan 22, 2020
The plant leucine-rich repeat receptor kinases GS O1/SGN3 and its peptide ligands CIF1 and CIF2 are essential for the formation of the Casparian strip. The Hothorn group from the Department of Botany and Plant Biology, in collaboration with the Geldner group from UNIL, has now uncovered in molecular detail how the SCHENGEN 3 receptor complex tightly binds CIF1 and CIF2.
O1/SGN3 and its peptide ligands CIF1 and CIF2 are essential for the formation of the Casparian strip. The Hothorn group from the Department of Botany and Plant Biology, in collaboration with the Geldner group from UNIL, has now uncovered in molecular detail how the SCHENGEN 3 receptor complex tightly binds CIF1 and CIF2.
Crystal structure of the GSO1/SGN3–CIF complex reveals a binding pocket for sulfotyrosine and extended back-bone interactions with CIF2. Structure-guided sequence analysis allowed to uncover previously uncharacterized CIF peptides conserved among higher plants. Quantitative binding assays with known and novel CIFs suggest that the homologous LRR-RKs GSO1/SGN3 and GSO2 have evolved unique peptide binding properties to control different developmental processes. A quantitative biochemical interaction screen, a CIF peptide antagonist and genetic analyses together implicate SERK proteins as essential coreceptor kinases required for GSO1/SGN3 and GSO2 receptor activation.
This work provides a mechanistic framework for the recognition of sequence-divergent peptide hormones in plants and was published in PNAS on January 21, 2020.
Michael Hothorn receives ERC consolidator grant
UNIGE press release, Dec 3, 2018
An ERC CoG has been awarded to Michael Hothorn for his research project entitled « System-wide discovery and analysis of inositol pyrophosphate signaling networks in plants (InsPire) ». The aim is to uncover nutrient signaling networks in plants. The grant is endowed with 2 million Euros over 5 years.
Philanthropies Select 41 Scientists as International Research Scholars
Inositol pyrophosphate sensor domains control phosphate homeostasis in eukaryotic cells
ESRF highlight 2016
SPX domains of previously unknown function are present in fungi, plants and animals. Several SPX domain crystal structures reveal a novel fold and a binding site for inositol pyrophosphates, enigmatic signalling molecules whose levels change in response to nutrient starvation.
SPX domains are small, soluble domains found in fungi, plants and animals. They can exist as stand-alone modules but are often located at the N-termini of proteins involved in phosphate uptake, transport, storage, metabolism or signalling. We determined 3.3 – 1.9 Å structures of fungal and human SPX domains from crystals obtained by carrier driven crystallisation and including data collected at beamline ID29 [1]. The different structures revealed a new fold with two long a-helices, connected by linkers of variable size. These core helices and two smaller C-terminal helices together form a 3-helix bundle, which is preceded by a flexible N-terminal helical hairpin (Figure 91). Many invariant lysine residues, which represent sequence fingerprints for SPX domains, are clustered in proximity to the N-terminal hairpin structure. A combination of genetic and biochemical experiments in yeast and Arabidopsis revealed that this basic surface represents a docking platform for inositol pyrohosphosphates (PP-InsPs), signalling molecules with poorly characterised cellular functions (Figure 91).
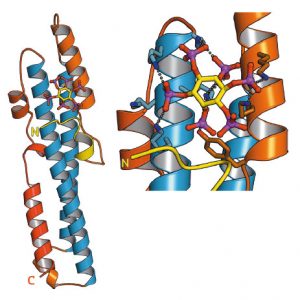
The concentrations of PP-InsP are known to change in cells, depending on whether they are supplied with sufficient amounts of inorganic phosphate or whether they experience phosphate starvation. We could demonstrate that SPX domains, when bound to PP-InsPs can interact with other proteins. In plants, one such interaction partner is a transcription factor, which is switched on under phosphate starvation to induce expression of genes that allow the plant to respond to the lack of this important nutrient. Under normal growth conditions, when PP-InsP levels are high, the transcription factor is kept inactive by forming a PP-InsP-dependent complex with a plant SPX domain. Under phosphate starvation, the lack of PP-InsP leads to dissociation of the transcription factor – SPX domain complex, thereby enabling the transcription factor to transcribe its target genes.
Taken together, our work defines SPX domains as cellular receptor for PP-InsPs, which control phosphate uptake, transport, storage, metabolism and signalling in fungi, plants and animals.
A cellular sensor of phosphate levels
UniGe press release April 14, 2016.Inorganic phosphate is an essential building block of cell membranes, DNA and proteins. It is also a main component of ATP, the “cell currency” of energy transfer. All cells therefore need to maintain a sufficient concentration of phosphate in their cytoplasm and have developed systems to transport and store this nutrient. But how does a cell know how much phosphate it actually needs? Researchers from the University of Geneva (UNIGE) and the University of Lausanne (UNIL), Switzerland, report that a region of specific proteins, the so-called SPX domain, signals the phosphate status to fungal, plant and human cells. This domain provides a binding surface for small molecules that regulates the uptake of the nutrient into the cell. Their findings, which now appear in Science, could contribute to the development of crops that use phosphate more efficiently.
In order to function properly, eukaryotic cells, i.e. cells from higher living organisms, need to maintain sufficient phosphate levels. To absorb this macronutrient, fungal, plant and human cells have developed transport and storage systems. How cells know how much phosphate they contain at any given time remained however unclear. Michael Hothorn, Professor at the Department of Botany and Plant Biology of the faculty of Science of UNIGE, and his research group revealed the crystal structure of a novel protein domain called SPX, which is involved in many phosphate signaling pathways. They discovered that SPX provides a binding surface for small compounds called inositol pyrophosphate signaling molecules (InsP), which can interact with other proteins only when they are bound to the SPX domain. As SPX domains can be attached to different proteins, such as enzymes, transporters or signaling proteins, the biologists hypothesized that InsP regulate various cell processes involved in phosphate homeostasis, from yeast to human cells.
A ubiquitous signal of the cell’s phosphate status
This hypothesis was explored in collaboration with researchers from the UNIL and other European universities. «We found out that the concentration of InsP changes in response to phosphate availability. InsP levels are high in cells that have sufficient phosphate, and drop when phosphate becomes scarce», explains Ruta Gerasimaite from UNIL, one of the first co-authors of the study. «In phosphate-starved plants, specific transcription factors turn on the expression of phosphate transporter genes. Once the plant is satiated, SPX domains filled with InsP will bind and inactivate these transcription factors, and no more phosphate will be absorbed from the soil into the cell», says Rebekka
Wild from UNIGE, another of the first co-authors. Yves Poirier, Professor at the Department of Plant Molecular Biology of UNIL, and his colleague Ji-Yul Jung further demonstrated this with the model plant Arabidopsis thaliana: when SPX domains present in phosphate transporters are mutated in the spot that normally binds InsP, phosphate transport is impaired. The role of InsP was initially elucidated in yeast cells: «We came across InsP while studying the mechanism of phosphate polymerization – its assembly into long chains – for the storage of this compound, and our data show that the SPX domain is a receptor for InsP», states Andreas Mayer, Professor at the Department of Biochemistry of UNIL. Once the SPX domain is filled, it activates the enzyme involved in phosphate storage.
Better understanding of phosphate homeostasis
This research now opens up new pathways to study and better understand phosphate homeostasis in organisms and may even lead to a more efficient phosphate use in crops. «Crops on the fields are usually lacking phosphate and therefore need to be fertilized. However, the resources of phosphate fertilizer are declining worldwide. Our discovery could open the door to the development of crops that could grow efficiently on less phosphate», says Michael Hothorn.
Unveiling the withering process
UniGe press release April 14, 2016.During their life, plants constantly renew themselves. They sprout new leaves in the spring and shed them in the fall. No longer needed, damaged or dead organs such as blossoms and leaves are also cast off by a process known as abscission. By doing so, plants conserve energy and prepare for the next step in their life cycle. But how does plant know when it is the right time to get rid of unnecessary organs? Researchers from the University of Geneva (UNIGE) and the University of Oslo (UiO) now shed light on this process. It is regulated by receptor proteinslocated at the surface of specific cells that form a layer around the future break point.
When it is time to shed an organ, a small hormone binds to this membrane receptor and, together with a helper protein, the abscission process is initiated. Their findings are now published in the journal eLife.
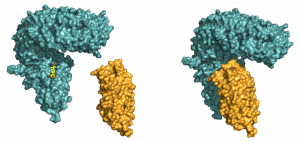
“It was already known that the membrane receptor protein HAESA and a small peptide –a short chain of amino acids – hormone called IDA are involved in the same signaling pathway and, together, control the shedding of floral organs. So far, however, the mechanism underlying their interaction was poorly understood”, explaind Michael Hothorn, professor at the Department of Botany and Plant Biology of the Faculty of Science of UNIGE. By solving the crystal structure of HAESA (from the Greek ‘to shed’) in complex with IDA, Hothorn and his team found out that the receptor directly senses the peptide hormone. They observed that HAESA contains a small cleft into which IDA fits perfectly. However, it only binds halfway to the receptor. To fully initiate the abscission process, another player is needed: the helper protein SERK1. IDA then works like a double-sided Scotch tape that tethers the entire complex together. The binding of SERK1 to HAESA and IDA triggers the molecular switch that instructs the cell to shed the organ.
Multitasking helper protein
“The fascinating thing about SERK1 is that it not only plays a role in the shedding mechanism of plant organs, but also acts together with other membrane receptors that regulate totally different aspects of plant development”, says Julia Santiago, first author of the study. Indeed, SERK1 is a versatile helper protein shared between different signaling pathways. When bound to another protein receptor, it can also for example signal the plantto grow. To verify their findings, the biologists from UNIGE collaborated with Melinka Butenko’s group from UiO. By studying genetically modified Arabidopsis thaliana plants, the Norwegian researchers confirmed the role of SERK1 in plant organ shedding. Their data were then statistically analyzed by professor Ludwig Hothorn from the Leibniz University Hannover. Now that they found out what is going on at the surface of plant cells before organ shedding, Michael Hothorn and his team want to discover what happens inside the cell. “ How exactly the molecular switch for abscission works is still largely unknown”, explains Michael Hothorn.
Twenty-seven researchers named as EMBO Young Investigators
EMBO press release November 12, 2014.EMBO announced today the selection of 27 young researchers as EMBO Young Investigators. The scientists join a network of 342 current and past Young Investigators who represent some of the best young group leaders contributing to research in Europe and beyond.
“The status of EMBO Young Investigator helps researchers under the age of 40 build their first independent teams and achieve a level of recognition that offers immediate benefits,” says Gerlind Wallon, Manager of the Young Investigator Programme. “The networking activities provide an additional layer of support for the young researchers.”
This year the programme received 202 applications. Thirteen per cent of the candidates who applied were selected and the new investigators originate from 11 countries.
The Young Investigator Programme provides support for researchers who have established their first laboratories in the past four years. The researchers receive a range of benefits, including an award of 15,000 Euros and the opportunity to apply for additional funds to help start their first independent research laboratories. Laboratory management and non-scientific skills training as well as PhD courses offer the young group leaders and their students the chance to develop professional skills. The scientists also receive access to core facilities at EMBL and funding for themselves and their group members to attend conferences.
Atomic insights into plant growth
Max Planck Society press release August 9, 2013. Source article Santiago, Henzler & Hothorn, Science, 2013If one wants to better understand how plants grow, one must analyze the chemistry of life in its molecular detail. Michael Hothorn from the Friedrich-Miescher-Laboratory of the Max Planck Society in Tübingen and his team are doing just that. Their latest work, published this week in Science, now reveals that a plant membrane receptor requires a helper protein to sense a growth-promoting steroid hormone and to transduce this signal across the cell membrane.
Every cell is surrounded by a greasy cell membrane. Signals from other cells and from the environment must be sensed at the cell surface, transduced across this membrane and translated into a specific response inside the cell. All organisms have evolved membrane receptor proteins to get these complex tasks done, but plant membrane receptors look drastically different from the well studied players in animals and bacteria. The plant steroid receptor BRI1, which can sense a small steroid hormone promoting plant growth, belongs to the family of leucine-rich repeat (LRR) receptor kinases, which are responsible for most membrane signaling events in plants. It was previously shown that BRI1 directly binds the small steroid hormone with its LRR-domain at the cell surface. Julia Santiago, a postdoctoral fellow in the Hothorn lab, could now demonstrate that BRI1 requires a helper protein to correctly sense the hormone and transduce the signal across the membrane. The helper SERK1 is a known player in the brassinosteroid signaling pathway, but it came as a surprise to see how early on it is required. By hitting protein crystals containing the ternary BRI1 – steroid hormone – SERK1 complex with intense X-rays, Santiago could see that SERK1 contributes directly to the formation of the hormone binding pocket, with both proteins interacting with the hormone. The steroid thus acts as a molecular glue which promotes association of the BRI1 and SERK1 LRR domains at the cell surface. This then causes interaction of the cytoplasmic kinases domains in the cell interior, which in turn activates a well characterized signaling pathway triggering the growth response.

The interesting feature of SERK1 is that it can help activate several seemingly unrelated plant receptor kinases, which bind vastly different ligands and trigger different responses. The new structures provide a first glimpse on how SERK1 might be able to do that. Instead of shaking hands with BRI1, it only uses a few ‘finger tips’ to contact the receptor. Other, strictly conserved surface patches remain available for the interaction with other plant receptor kinases and, potentially their ligands. “There must be some advantage to having all this different functions combined into a single helper protein”, Hothorn speculates. Notably, the use of a shared helper protein could allow different signaling pathway to communicate with each other.
The atomic models offer other novel insights too: “Looking at our models, we can now predict pretty well, which mutation in the receptor or helper protein should have an effect on the down-stream signaling pathway. We also know what parts of the hormone are really important to make it bind to the receptor or to the helper protein”. Such detailed insights may promote the rational design of synthetic plant steroid hormones and receptor antagonist with applications in basic research, and perhaps someday in the field.
Three new ERC Grants for young Tübingen Max Planck scientists
Max Planck Society press release September 18, 2012.Fulvia Bono, research group leader at the Max Planck Institute for Developmental Biology, as well as Wolfram Antonin and Michael Hothorn, both Max Planck research group leaders at the Friedrich Miescher Laboratory in Tübingen, will each be awarded a Starting Grant of the European Research Council (ERC). For the realization of their project ideas evaluated as scientifically excellent, the three scientists will each receive up to 1.5 million Euros over the next five years. ERC Starting Grants aim to support up-and-coming research leaders at an early stage of their career to conduct independent research in Europe.
According to the ERC, 800 million Euros will be spend to award 536 researchers in Europe Starting Grants this year. 4,741 scientists Europe-wide had applied for the highly funded grants that are awarded for scientifically excellent research proposals. ERC Starting Grants represent a personal distinction for the individual scientist and provide funding for up to 5 years. This year’s awards highlight the scientific excellence of the entire Tübingen Max Planck Campus: Including 2010 ERC Starting Grant recipients Gáspár Jékely and Richard Neher from the Max Planck Institute for Developmental Biology, five of the 13 independent young research group leaders at the MPI for Developmental Biology and the Friedrich Miescher Laboratory are now funded by ERC Starting Grants.

Since 2011, Fulvia Bono has been leading an independent research group at the Max Planck Institute for Developmental Biology. She investigates the role of the intracellular transport of macromolecules in the regulation of gene expression. With the funds from the ERC Starting Grant she will extend her research to include the study of the crucial role that the correct messenger RNA (mRNA) localization in the cell cytoplasm plays during animal development, in the maintenance of cell polarity and in nervous system function.
Messenger RNA is combined with certain proteins to form particles, the so-called mRNP complexes. In the fruit fly Drosophila, the localization of these complexes determines the formation of the embryo’s body axis. Fulvia Bono wants to understand the function of the mRNP systems at a mechanistic level and gain deeper insight into the connection between genes and outer appearance of organisms.
With the ERC Grant she will recruit several new members for her research group. “With the expanded resources of the ERC grant, I can accept some more scientific challenges I could not have thought of otherwise,” she says. Fulvia Bono received her PhD from the University of Pavia in 2000 and continued her research at the European Molecular Biology Laboratory (EMBL) in Heidelberg for five years. From 2008 to 2011, she was a project leader at the Max Planck Institute for Developmental Biology.
Wolfram Antonin has been a Max Planck research group leader at the Friedrich Miescher Laboratory since 2006. His main research topic is the breakdown of the nuclear envelope, which separates the nucleus with the DNA from cell cytoplasm, in the process of cell division and its reassembly after formation of the daughter cells. With the ERC Starting Grant he will be able to start an additional project with several new team members and pursue his interest in DNA decondensation.
During cell division, the DNA has to be contracted up to 50 times in the chromosomes to a transport form. After the formation of the new daughter cells, the DNA is unpacked again for transcription and replication. Wolfram Antonin had noticed that, while the packing of the DNA is a well-studied process, little is known of DNA decondensation. “It is possible,” the scientist says, “that DNA decondensation is a passive process, as if a clip around the DNA thread is released and the DNA just relaxes.” However, initial experiments hint at an active process. Wolfram Antonin wants to investigate this process and the proteins involved.
“This is a high-risk research project, since we cannot say anything about the outcome. This is special about the ERC grants, that the sponsor trusts the skills of the scientists themselves and funds promising project ideas,” he says. Wolfram Antonin obtained his PhD at the Max Planck Institute for Biophysical Chemistry in Göttingen in 2001. After receiving post-doctoral training at the European Molecular Biology Laboratory (EMBL) in Heidelberg, he joined the Friedrich Miescher Laboratory in 2006.
Since the beginning of 2012, Michael Hothorn has been building up his Max Planck research group at the Friedrich Miescher Laboratory. His research field is structural plant biology; he investigates signaling pathways in plant cells. With the funding from the ERC Starting Grant he will be able to start an additional project on the search for the enzyme responsible for the assembly of phosphate polymers in plant cells. The corresponding enzyme in bacteria has been known for a long time, yet it does not exist in the cells of higher organized organisms, like plants or mammals. “The function of the phosphate polymers is enigmatic,” the scientist says. “It has been suggested that they are used to store phosphate in plant cells and tissues.” The project may, in the future, have applications in crop science, as, at present, many crop species require the application of phosphate fertilizers.
“Without the ERC Grant, I could not afford a project of this size and kind. The sparse results from our first experiments would not have been sufficient for the usual ways of obtaining research funding. The prospect of our high-risk project is to find out something fundamentally new, yet the risk is to receive detached data we are not able to connect,” the scientist says. After obtaining his PhD at the European Molecular Biology Laboratory (EMBL) in Heidelberg in 2006, Michael Hothorn carried out research at the Salk Institute for Biological Studies in La Jolla before joining the Friedrich Miescher Laboratory in 2011.
Michael Hothorn wins HFSP career development award
Max Planck Society press release March 29, 2012.Michael Hothorn, biologist and Max Planck research group leader at the Friedrich Miescher Laboratory of the Max Planck Society in Tübingen, receives a Career Development Award from the Human Frontier Science Program (HFSP) for a basic research project in the field of structural plant biology. The three year project will start in August 2012 and is funded with 300.000 US Dollar.
Michael Hothorn investigates receptor kinases, membrane proteins located at the cell surface of plant cells. These molecules act as signal transmitters in many important processes like growth and development, in immune defense responses or in the initiation of symbiotic interactions. “Receptor kinases are able to recognize specific substances on the cell surface. When these ligands bind, a kinase module inside the cell is switched on,” the scientist explains. Kinases are enzymes, small catalysts that drive certain chemical reactions. Michael Hothorn wants to know what happens in the short time span between ligand binding on the cell surface and the onset of the kinase reaction. Over two-hundred such receptors exist in the small genome of Arabidopsis thaliana, an important plant model organism.
Career Development Awards are granted to selected HFSP Long-Term Fellows who have done postdoctoral research abroad and return to their home country. The Award supports scientific independence of young researchers and helps them to establish their own laboratory. Michael Hothorn graduated from the University of Heidelberg and received his PhD from the European Molecular Biology Laboratory (EMBL). From 2008 to 2011, he was a HFSP Long-Term Fellow at the Salk Institute for Biological Studies in San Diego, California. Since January 2012 he is leader of the junior research group Structural Plant Biology at the Friedrich Miescher Laboratory in Tübingen.
Participants of the HFSP are Australia, Canada, India, Japan, Republic of Korea, Norway, New Zealand, Switzerland the United Kingdom, the United States of America and the European Union. The HFSP supports innovative and interdisciplinary basic research focused on the complex mechanisms of living organisms. The Career Development Award is conferred to applicants proposing an innovative frontier research program that holds promise for the development of new approaches to problems in the life sciences with potential to advance the field of research significantly. Michael Hothorn is one of eight awardees of 2012 who, according to the HFSP, were selected from 55 applicants.
Plant hormone receptor inherited from the bacterial world
HFSP press release October 3, 2011. Source article: Hothorn, Dabi & Chory, Nat Chem Biol, 2011Plants, just like animals, use small molecule hormones to integrate the growth and development of their cells and tissues. The hormone receptor proteins that recognize these ligands are however very different in plants and animals. A new study now confirms that plants use signaling proteins normally found in bacteria to initiate certain hormone responses.
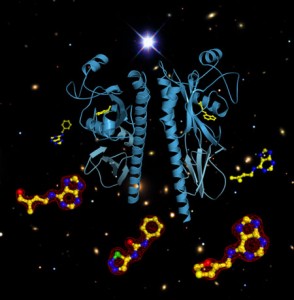
One of the most fundamental tasks of a cell is to recognize signals from the outside world at the cell surface, and to relay these signals to the cell’s interior. The different kingdoms of life have evolved different membrane receptor proteins to fulfill this task. Interestingly, plant genomes contain receptors that are closely related to the animal world, but also proteins that are somewhat reminiscent of sensor histidine kinases, the key players in bacterial membrane signaling. It has been debated whether these proteins were indeed adopted from the bacterial world, or whether they evolved independently. One such receptor is the Arabidopsis histidine kinase 4 (AHK4), a membrane-integral receptor for the classic plant hormone cytokinin. A crystal structure of its extracellular hormone binding domain now reveals just how closely related plant cytokinin receptors are to bacterial signaling proteins.
Hothorn et al. solved the structure of AHK4’s hormone binding domain mainly to understand how this receptor can interact with chemically very diverse cytokinin ligands, natural cytokinins and synthetic cytokinins which are important growth regulators and defoliants in agriculture. The structures, however, did not only reveal how the receptor can accommodate these different molecules: AHK4 turned out to look exactly like a bacterial signaling protein, suggesting that the plant receptor may have originated from an bacterial ancestor. And there was a ligand stuck in AHK4s hormone binding pocket that looked exactly like a plant cytokinin, but was carried over from the engineered bacterial expression strain used to produce AHK4 for structural studies. This meant that bacteria did not only contain signaling proteins with a shape very similar to AHK4, but also small molecule ligands that look exactly like the plant hormone. These findings allow speculation that cytokinin signaling may have evolved already in bacteria, and was later adopted to control growth, development and the integration of stem cell populations in higher plants.
On the more practical side, the different AHK4 structures suggest how to design new cytokinins rationally, with potential application in basic research and in the field.
Study Reveals Important Aspects of Signalling Across Cell Membranes in Plants
NSF press release June 13, 2011. Source article: Hothorn et al., Nature, 2011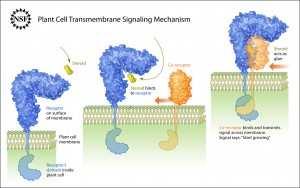
Credit: Zina Deretsky, National Science Foundation
Every living plant cell and animal cell is surrounded by a membrane. These cellular membranes contain receptor molecules that serve as the cell’s eyes and ears, and help it communicate with other cells and with the outside world.
The receptor molecules accomplish three basic things in the communication process: 1) recognize an outside signal, 2) transport that signal across the cell’s membrane and 3) initiate the reading of the signal inside the cell and then initiate the cell’s response to that signal. These steps are collectively known as transmembrane signaling.
Transmembrane signaling in animal cells has been significantly more studied and observed than that in plant cells. But now, with support from the National Science Foundation, researchers from Joanne Chory’s laboratory at the Salk Institute have published new observations about transmembrane signaling in plants; their paper appears in the June 12, 2011, advanced online edition of Nature.
According to the study, transmembrane signaling mechanisms used by plants differ from those used by animals. Specifically, Michael Hothorn of the Salk Institute reports that a small steroid molecule on the outside of the plant cell assists in the transmembrane signaling process. By contrast, this sort of molecule and its receptor is generally located inside the nuclei of animal cells.
While studying transmembrane signaling in plants, Hothorn and colleagues observed the steroid, shown in yellow, attach to a membrane-bound receptor, shown in blue. This attachment enabled the steroid’s counterpart–a co-receptor protein, shown in orange–to bind to the blue receptor. Once bound, the orange co-receptor and the blue receptor become glued together by the yellow steroid, allowing their intracellular domains to touch and initiate communication.
In the case observed by Hothorn, transmembrane signaling initiated plant growth.
Plant receptors reflect different solutions for signaling problem
Salk press release June 13, 2011. Source article: Hothorn et al., Nature, 2011La Jolla—Birds do it, bees do it, and for most things biological, even plants do it. But not necessarily like their animal counterparts. A study led by Salk Institute scientists shows that a plant receptor does one of the most fundamental cellular “its”—the delivery of a hormonal signal from outside the cell to the nucleus—in a radically different way than its animal cousins. Knowing that could aid creation of techniques to speed plant growth and enhance agricultural production.
In a study published in the June 12, 2011, advance online edition of the journal Nature, a team led by Joanne Chory, Ph.D., professor and director of the Plant Molecular and Cellular Biology Laboratory and a Howard Hughes Medical Institute investigator, reports the three-dimensional structure of a plant steroid hormone receptor known as BRI1.
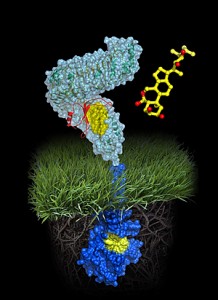
A molecule of brassinolide (yellow wire model) binds to the extracellular domain of the receptor (in light-blue). Binding ultimately causes phosphorylation of the receptor’s cytoplasmic kinase domain (in dark blue), thereby transducing the signal across the membrane.
Image: Courtesy of Michael Hothorn and Jamie Simon, Salk Institute for Biological Studies
Since the late 90’s the Chory lab has conducted landmark studies revealing that plant and animal steroids are made via similar pathways and that, like animals, plants use steroids to become bigger, regulate sexual development, and control physiology.
The new study caps that work with a molecular analysis of a receptor that transduces the plant steroid signal. “Our genetics studies previously showed that unlike animal steroid receptors, which bind steroids inside cells, plant steroid receptors are membrane proteins, a completely different class of protein,” says Chory, holder of the Howard H. and Maryam R. Newman Chair in Plant Biology. “Now that we know the precise contacts made between the steroid and its receptor, we can propose how the BRI1 receptor works.”
The new work reports the atomic structure of BRI1 and then superimposes it with a distant structural cousin, the mammalian membrane protein TLR3, which activates an innate immune response in mice and humans. The TLR3 structure was determined in 2005 by Ian Wilson, Ph.D., professor in the Department of Molecular Biology and member of the Skaggs Institute at The Scripps Research Institute (TSRI) and a co-author of the current study.
The comparison affirms that, although somewhat similar to TLRs architecturally, the domain of BRI1 that pokes out through the cell membrane and monitors the extracellular environment exhibits twists and turns unique to plants. “We thought BRI1 would look like TLR3, which is shaped like a horseshoe,” says Michael Hothorn, Ph.D., a postdoctoral fellow in the Chory lab and the study’s first author. “But instead BRI1 was twisted into a superhelical spiral.”
To visualize those twists, the group employed a technique called x-ray diffraction. That method requires that scientists first grow highly purified crystals of BRI1 extracellular “antenna”—in this case derived from the mustard plant Arabidopsis thaliana—and then bombard the crystals with x-rays. The way x-rays bounce off, or “diffract” from, the crystal enables researchers to construct a three-dimensional, Lego-like representation of the protein’s architecture in the presence or absence of steroid activator.
One prediction was that structural shifts caused by binding of the brassinolide steroid might resemble conformation changes made by TLR when it initiates an immune response. “We knew that when the TLR3 horseshoe binds an activator another horseshoe becomes glued on top of it,” says Hothorn. “But BRI1 contains an island domain that first binds steroid onto the twisted structure and then provides a platform for a different protein to interact and relay the signal.”
Although the job of proteins like TLR3 and BRI1 is to alter gene expression patterns in response to environmental stimuli, their structural differences likely reflect the fact that those stimuli are fundamentally different molecules. “In mammals TLR3 is an innate immunity receptor activated when large ligands such as viral RNA bind to a highly repeated area called the LRR or Leucine-Rich Repeat domain,” says Wilson. “BRI1 is also an LRR protein but its structure is highly specialized to recognize and respond to smaller plant steroid hormones.”
“Michael’s structural work is the final brick in the wall,” says Chory, noting that BRI1 serves as the prototype for a large class of similar proteins expressed in plants. Interestingly, BRI1 is an exception in that family: while its job is to relay growth-promoting signals, many of its look-alikes actually stimulate immune responses in plants, protecting them from insects, worms or bacteria. Whether BRI1’s sibling receptors display such a twisted structure opens new avenue of investigation.
Many common herbicides were designed to mimic the structure of plant hormones. “Because brassinosteroids are hormones, knowing the structure of their receptor will allow us to rationally design herbicides that could block interaction between hormone and receptor,” says Chory. “This would enable us to manipulate how fast plants grow and how large they become-traits that are important in crops that must soon feed 10 billion people.”
Also contributing to the work were Youssef Belkhadir and Tsegaye Dabi of the Chory lab, Joseph Noel of Salk, and Marlene Dreux of The Scripps Research Institute in La Jolla.
Support for the work was from the Howard Hughes Medical Institute, the National Science Foundation, the European Molecular Biology Organization, the International Human Frontier Science Program Organization, the Philippe Foundation, the National Institutes of Health, and the Skaggs Institute for Chemical Biology at TSRI.
Tyrosine phosphorylation regulates plant growth
HFSP press release March 31, 2011, Source article: Jaillais et al., Genes Dev, 2011The plant steroid hormone receptor BRI1 is a membrane-bound receptor kinase that controls an important growth signaling pathway in plants. Jaillais et al. now report that BRI1 uses tyrosine phosphorylation, an on/off switch that was thought to be restricted to the animal kingdom, to dissociate its own inhibitor protein from the plasma membrane.
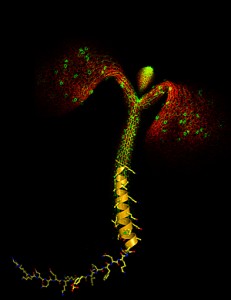
Image: Courtesy of Yvon Jaillard, Michael Hothorn and Jamie Simon, Salk Institute for Biological Studies. Image appeared on the cover of Genes & Development vol 25(3), Feb 1 2011.
Signal perception at the cell surface and transduction of this signal to the cell’s interior is essential to all life forms. Animals mainly use G-protein coupled receptors and tyrosine receptor kinases to perceive such signals. Plants however evolved membrane-integral receptor kinases that are very different from the receptors found in animals and usually have serine/threonine kinase activity. A well studied family member is the brassinosteroid receptor BRI1, which controls an essential growth signaling pathway. BRI1 is kept in its basal state by its intracellular tail and by interaction with a small inhibitory protein known as BKI1. Autophosphorylation of the BRI1 receptor is necessary to release the inhibitory protein into the cytosol and to activate the steroid signaling pathway, but the molecular mechanism has been enigmatic.
Jaillais et al. have now discovered that BKI1 acts through two evolutionarily conserved motifs: a 20-amino-acid sequence that binds the receptor kinase domain and a lysine-arginine-rich motif that anchors the inhibitory peptide to the plasma membrane. BRI1-mediated phosphorylation of a key tyrosine within the membrane-targeting motif releases BKI1 from the membrane, relieving kinase inhibition and allowing for the formation of an active signaling complex.
The phosphorylation of BKI1 is the first documented example of tyrosine transphosphorylation in plants, and importantly, the phosphorylation and release of kinase inhibitor proteins appears a unifying theme in tyrosine kinase signaling in animals and plants.
By defining common features in plant and animal receptor signaling pathways, the Salk researchers hope to learn more about what the requirements for a robust signaling system are. Although plants don’t encode canonical tyrosine kinases in their genomes, the authors predict that tyrosine phosphorylation will emerge as an important topic in plant signaling.
New study reveals the protein that makes phosphate chains in yeast
It can be found in all life forms, and serves a multitude of purposes, from energy storage to stress response to bone calcification. This molecular jack-of-all trades is polyphosphate, a long chain of phosphate molecules. Researchers at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, are now the first to uncover how this chain is assembled in eukaryotes (organisms whose cells have a nucleus). The study, published this week in Science, uncovers the function of a single protein with a wide range of potential implications ranging from improving crops to fighting diseases such as sleeping sickness.
Scientists have known for a long time how bacteria make phosphate chains, but how the same process works in eukaryotes has so far remained elusive. EMBL scientists now show that in yeast a protein called Vtc4p is responsible for the production of polyphosphates. Vtc4p is part of a protein complex called vacuolar transporter chaperone complex (VTC) that is usually found in the membranes of vacuoles – pouches in which cells store molecules for later use, transport or destruction.
“This protein is like a factory,” says Klaus Scheffzek, whose group carried out the research at EMBL in collaboration with the Département de Biochimie at the Université de Lausanne, Switzerland, and others,”it sits in the vacuolar membrane, generates long chains of polyphosphates and we speculate that it sends them straight to the vacuole for storage.”
Vtc4p is partly embedded in the membrane and has a ‘tail’ hanging into the cell, which removes a phosphate molecule from ATP, an important energy carrier in the cell. Vtc4p uses the energy that is released by that cleavage to add the newly acquired phosphate to a growing chain of phosphates. Since the rest of Vtc4 straddles the membrane, scientists suspect this protein probably transfers the polyphosphate chain to the vacuole as it produces it.
The researchers determined Vtc4p’s function by looking at its 3D structure.
“This study emphasises the importance of structural biology not just to show what molecules look like and how they work but also what that function is,” says Michael Hothorn from Scheffzek’s group at EMBL, who is presently at The Salk Institute for Biological Studies in California.
Since polyphosphate is a ubiquitous, multi-tasking molecule with many different functions, discovering how it is produced could have implications for many different fields. Although Vtc4p is not present in plants, the discovery could have implications for agriculture, for instance in the production of fertilizers and high-yield crops. Polyphosphate is important for plant growth, and the scientists suspect Vtc4p could play an important role in making it available to plants that have fungi living in their roots. Because the VTC can move from the membrane of the vacuole to that of the cell, it could assemble phosphate chains and transfer them to outside the fungus cell, where they would be available to the plant.
The research could also pave the way for new treatments for diseases such as sleeping sickness and Chagas disease, as the parasites that cause them need polyphosphate chains to survive.

